 » Barrier-free View
» Barrier-free View
| ||||||||
 | ||||||||
 |
 |
 |
| Picture of the 1st Network Meeting of the Herz-Kreislauf-NetzDownloadsArchivExternal job opportunities |
 |
Prevalence of Titin-mutations and identification of novel disease genes in patients with familial dilated cardiomyopathy
 |
Subproject leader
PD Dr. Dieter Weichenhan
University of Heidelberg
Internal Medicine III / Cardiology
Otto-Meyerhof-Center
Im Neuenheimer Feld 350
69120 Heidelberg
email: dieter.weichenhan@med.uni-heidelberg.de
Phone: 06221-56 1505
Dilated cardiomyopathy (DCM) is a complex myocardial disease characterized by unexplained dilation of the left ventricle and systolic dysfunction. The disease may cause sudden cardiac death or progressive heart failure that may eventually require heart transplantation. Familial aggregation of DCM is observed in 25-40% of cases (1), usually displaying an autosomal dominant mode of inheritance. More than 20 disease loci and candidate genes have been identified so far, mostly involving those for nuclear and cytoplasmatic cytoskeletal proteins as well as components of the sarcomeric Z-disc and the dystroglycan complex including lamin, desmin, muscle LIM protein, CYPHER/ZASP and δ-sarcoglycan (2, 3). In clinical practice, however, the genetic causes are only very rarely disclosed and used for prognosis of the disease. Moreover, the unusually high genetic heterogeneity makes the screening for DCM causing mutations to a search for the needle in a hay stack. Whereas large scale mutation screening is performed with great efforts in patients with hypertrophic cardiomyopathy (HCM) (e.g., http://cardiogenomics.org), mutation screening in patients with DCM is usually limited to single or only a few candidate genes and hence, relatively inefficient. Comprehensive genetic information for counseling patients and their relatives requires, however, extensive and reliable mutation screening which, in turn, may also provide valuable information on novel pathways associated with the disease and the functional relevance of protein domains and interaction sites.
To improve our still rather limited knowledge on the genetic and molecular causes of DCM, we collect and investigate idiopathic and familial cases of DCM in collaboration with partners from the NGFN cardiovascular disease net and other cardiological centers. Patients are screened for mutations in more than 20 candidate genes by denaturing gradient gel electrophoresis (DGGE). In a pilot study, we have already identified a considerable number of relevant known and novel mutations as well as many coding and non-coding polymorphisms.
We have established a large scale platform (Fig. 1) for mutation screening in the coding exons of 10 known and 16 novel candidate genes for DCM. Novel candidates do either encode components of essential structures of the cardiomyocyte, are aberrantly expressed in disease or involved in animal models for DCM (Table 1). Mutation screening is based on heteroduplex analysis by DGGE, a robust and highly reliable method (4), suitable for high-throughput analysis and enabling us to perform more than 500 analyses daily (Fig. 2). From 325 conding exons which represent the 26 genes, we presently analyse 305 (94%) by DGGE while the remainder may be analysed by direct sequencing.
Screening of 20 DCM and 30 HCM patients for mutations in genes MYBPC3, MYH7 and TNNT2, which are known candidates for both DCM and HCM, revealed 15 mutations in the HCM and 3 in the DCM patients (Table 2). Several of the mutations were novel. Additionally, we identified a large number of different coding and non-coding polymorphisms. Screening of the DCM patients alone for mutations in the remaining 23 genes identified one mutation each in genes LMNA, DMN, FLT1 and PXN. The latter three belong to the novel candidates and, hence, are of particular interest for further studies. In several instances, when blood from affected and unaffected family members of index patients was available, we observed a clear cut segregation of the mutation with the disease. These data underscore the causative nature of the mutation and may be of particular value in counseling affected families. Genotype and phenotype data are correlated in a database.
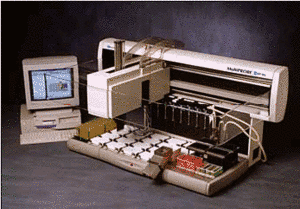 |
Semiautomatic DNA isolation from blood |
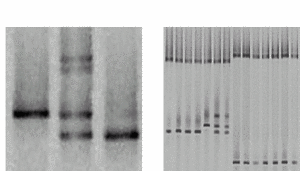 |
PCR of exons and DGGE analysis |
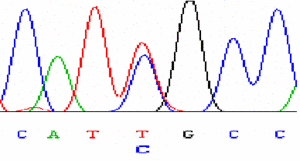 |
Sequencing of variants |
 |
Functional and structural studies of mutations Fig. 1: Workflow of the mutation screening platform |
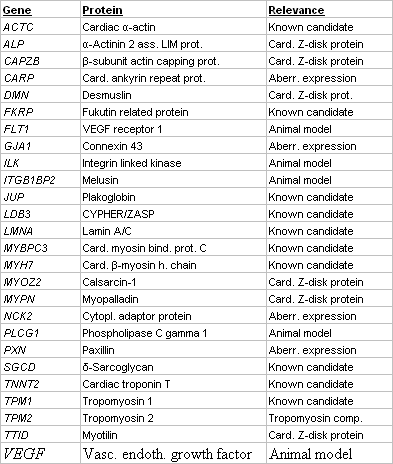 |
Table 1 Candidate genes for DCM screened by DGGE |
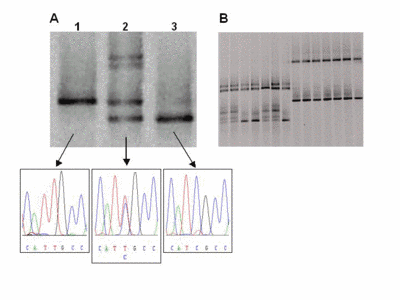 |
Fig 2: Representative DGGE results A) Lane 1 shows the single DNA band of a wildtype sample with a T/A base pair (arrow). In lane 2, four bands indicate a heterozygous patient. The upper part shows two bands from heteroduplexes formed by mismatches (C/A and T/G) in addition to the bands of the wildtype and the mutated (C/G base pair) allele. Lane 3 shows a homozygous mutant. B) Several samples can be analysed reliably in a single lane (multiplexing). |
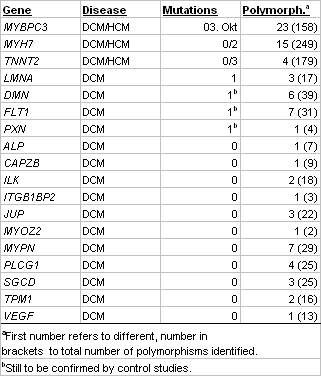 |
Table 2 Identified mutations and polymorphisms in pilot study |
This project aims to determine the prevalence of known and novel genes associated with familial DCM. Despite the relatively small number of patients screened so far, we have already identified a considerable number of mutations and polymorphisms in known and novel candidate genes, underscoring the efficiency of the screening platform. In collaboration with other NGFN members, novel mutations, particularly those in novel candidate genes, will be further analysed functionally in cellular assays and structurally by computer modeling. Patient recruitment and mutation screening will be continued and further extended by inclusion of additional candidate genes. Regular clinical follow-up of patients will enable us to correlate the time course of the disease with a particular genetic defect and, thus, provide novel insights in the prognosis of the disease.
Our screening platform is also open to other partners on a collaborative basis.
 |
Working group PD Dr. Weichenhan and Dr. Ivandic |
In cooperation with Prof. Dr. L. Thierfelder,
Max-Delbrück-Centrum of Molekulare Medicine, Berlin,
Prof. Dr. V. Regitz-Zagrosek, German Heart Center, Berlin
and Dr. P. Ehlermann, University of Heidelberg,
Internal Medicine III, Cardiology
Publications:
1. Grunig, E., Tasman, J.A., Kucherer, H., Franz, W., Kubler, W., and Katus, H.A. 1998. Frequency and phenotypes of familial dilated cardiomyopathy. J Am Coll Cardiol 31:186-194.
2. Seidman, J.G., and Seidman, C. 2001. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell 104:557-567.
3. Pyle, W.G., and Solaro, R.J. 2004. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ Res 94:296-305.
4. Nollau, P., and Wagener, C. 1997. Methods for detection of point mutations: performance and quality assessment. IFCC Scientific Division, Committee on Molecular Biology Techniques. Clin Chem 43:1114-1128.
more information about the working group
http://www.klinikum.uni-heidelberg.de/index.php?id=4573