 » Barrier-free View
» Barrier-free View
| ||||||||
 | ||||||||
 |
 |
 |
| Picture of the 1st Network Meeting of the Herz-Kreislauf-NetzDownloadsArchivExternal job opportunities |
 |
Antisense oligonucleotide-mediated gene knock-down in Zebra fish embryo for the evaluation of novel cardiovascular genes
 |
Subproject leader
Dr. Wolfgang Rottbauer
University of Heidelberg
Internal Medicine III / Cardiology
Otto-Meyerhof-Center
Im Neuenheimer Feld 350
69120 Heidelberg
email: wolfgang.rottbauer@med.uni-heidelberg.de
Phone: 06221-56 4697
The goal of the proposed research is to systematically characterize the in vivo function of novel cardiovascular genes. During NGFN-1, numerous expression studies on heart tissue from patients and animal models with different cardiovascular diseases revealed several hundred differentially regulated novel genes (see work of collaborators and own group). Their in vivo role in cardiovascular function and disease development now needs to be evaluated systematically to help reconstructing disease specific molecular pathways.
Mouse gene knock-out strategies, however, are time consuming and require huge financial resources. Very often, mice, homozygous for mutations in cardiovascular genes, die early during embryonic development due to loss of blood circulation, not allowing for proper characterization of the gene’s “loss of function” effects in the cardiovascular system. In contrast, zebrafish (ZF) embryos live by diffusion for several days, and thus heart function is largely dispensable during embryonic development and early larval live. The transparency of ZF embryos facilitates the assessment of embryonic cardiovascular function by light microscopy, with quantitation possible by direct hemodynamic measurements. The cardiovascular system develops extremely fast in the ZF embryo: Only 72 hours after fertilization (hpf) of the egg, cardiovascular morphogenesis is completed and the heart functions as a mature organ in the ZF larvae. Recently, injection of Morpholino-modified antisense oligonucleotides evolved as a fast and inexpensive method to study cardiovascular gene loss-of function phenotypes in the living ZF (Figure 1). The antisense oligonucleotides can be easily injected into ZF embryos, reliably inhibiting translation of the targeted gene for at least 96 hpf. Due to these circumstances the ZF evolved as a powerful genetic model organism to evaluate novel genes essential for the development and maintenance of vertebrate cardiovascular function.
 |
Fig. 1 Microinjection (A) of Morpholino antisense oligonucleotides against the zebrafish pontin gene induces cardiac hyperplasia (C) and heart failure. |
Using antisense oligonucleotide-mediated gene knock-down and cardiac-specific overexpression strategies in ZF, the proposed research aims to characterize about 250 novel genes, which were found to be differentially regulated in various cardiovascular diseases (Figure 2)
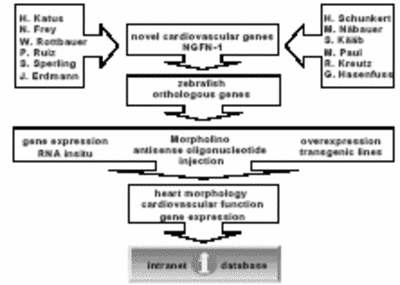 |
Fig. 2 Work flow of the zebrafish platform from candidate gene to the intranet data base. |
To do so, we first established in cooperation with Dr. Merk (IBE, Munich) a web-based database that allows for proper storage and accessibility of the acquired datasets including, images and videos of zebrafish embryos. According to our aims, we evaluated so far more than 50 novel genes that were found to be differentially regulated in various cardiovascular diseases for their function in the zebrafish cardiovascular system by antisense mediated gene-knock-down. To do so, corresponding ZF orthologous genes were identified, Morpholino-modified antisense oligonucleotides against these genes designed and into 1-8-cell-stage embryos injected. The effect of the gene knockdown on morphology and function of the cardiovascular system was characterized using digital video microscopy. Fractional shortening of the atrial and ventricular chamber as well as blood velocities were measured and electrocardiograms (ECGs) recorded when cardiac arrhythmias were found to be induced. Heart morphology was evaluated by light microscopy and digital imaging as well as structural and ultrastructural analyses performed. Timing and location of targeted gene expression using gene-specific antisense RNA probes was determined. It is planned to establish for a subset of genes cell-autonomous cardiac overexpression phenotypes (genetic mosaics) as well as heart-specific transgenic ZF lines. The mosaic and transgenic lines will be further characterized as outlined above.
The results of these experiments will reveal new gene functions and genetic pathways underlying cardiovascular diseases and will guide the design of proper transgenic animal models for further characterization of the gene’s functions in the mammalian heart (see Figure 2).
Publications:
1. Rottbauer W, Just S, Wessels G, Trano N, Most P, Katus HA, Fishman MC. VEGF-PLCgamma1 pathway controls cardiac contractility in the embryonic heart. Genes Dec 2005, 19:1624-1634
2. Blessing E, Rottbauer W, Mereles D, Hosch W, Benz A, Friess H, Autschbach F, Müller M, Stremmel W, Katus H. (2005). Isolated Left Ventricular Noncompaction of the Myocardium as a Cause of Embolic Superior Mesenteric Artery Occlusion. J Am Soc Echocardiogr 18: 5-7.
3. Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. (2005) High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 1 (5): 263-264
4. Rottbauer, W., Baker, K., Wo, Z.G., Mohideen, M.A., Cantiello, H.F., and Fishman, M.C. 2001. Growth and function of the embryonic heart depend upon the cardiac- specific L-type calcium channel alpha1 subunit. Dev Cell 1:265-275.
5. Xu, X., Meiler, S.E., Zhong, T.P., Mohideen, M., Crossley, D.A., Burggren, W.W., and Fishman, M.C. 2002. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet 30:205-209.
6. Rottbauer, W., Saurin, A.J., Lickert, H., Shen, X., Burns, C.G., Wo, Z.G., Kemler, R., Kingston, R., Wu, C., and Fishman, M. 2002. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell 111:661-672.
7. Nasevicius, A., and Ekker, S.C. 2000. Effective targeted gene 'knockdown' in zebrafish. Nat Genet 26:216-220.
more information about the working group:
http://www.klinikum.uni-heidelberg.de/index.php?id=4580