Genetic epidemiology of heart failure
 |
Subproject leader
Prof. Dr. Heike Bickeböller
Department of genetic Epidemiology
Georg-August-University of Göttingen
Humboldtallee 32
37073 Göttingen
email: hbickeb@gwdg.de
Phone: 0551-39 14020
The aim of the subproject Genetic Epidemiology of Heart Failure is to support the Heart Failure Projects regarding biostatistics and genetic epidemiology in their endeavour for the identification, quantification and functional investigation of susceptibility genes. We participate in all project phases, i.e. study planning (study type, power, sample size) and analysis (statistical analysis, interpretation, communication of results).
Study planning must account for methods in biostatistics, bioinformatics (e.g. association methods). Interdisciplinary work is necessary between clinicians, core centres and genetic epidemiology for analysis and result communication as well.
The aim of the here reported studies are two-fold: i) We wanted to assess the impact of the individual genetic makeup on doxorubicin-induced cardiotoxicity in a case-control-study. Identification of the involved genes and their variants could help to develop safer individualized therapies and also give further insight into the pathophysiology of this and other types of heart failure. ii) We wanted to evaluate new technologies such as genome-wide association genotyping and genomic controls. The case-control study and the evaluation of genome-wide association genotyping have been carried out as interdisciplinary work under the leadership of Prof. Wojnowski, University of Mainz (Project Genomic Predictors of Heart Failure Following Anthracycline Cancer Therapy).
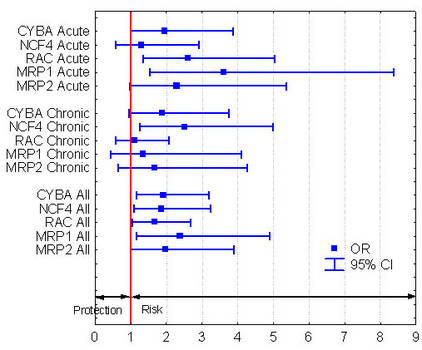
We looked for genetic markers predictive for doxorubicin-induced heart failure in patients with Non-Hodgkin lymphoma of the German Non-Hodgkin Lymphoma (NHL-B) study who were followed up for the development of heart failure for a median of more than 3 years. SNPs were selected from 82 genes with conceivable relevance to ACT. Out of 1697 patients, 55 developed an acute and 54 a chronic ACT (cumulative incidence of either form 3.2%). We detected 5 significant associations with polymorphisms of the NAD(P)H oxidase and doxorubicin efflux transporters. Chronic ACT was associated with a variant of the NAD(P)H oxidase subunit NCF4 (rs1883112, -212A>G, OR: 2.5, 95% CI: 1.3 - 5.0). Acute ACT was associated with the His72Tyr polymorphism in the p22phox subunit (rs4673, OR: 2.0, 95% CI: 1.0 - 3.9) and with the variant 7508T>A (rs13058338, OR: 2.6, 95% CI: 1.3 – 5.1) of the RAC2 subunit of the same enzyme. In addition, acute ACT was associated with the Gly671Val variant of the doxorubicin efflux transporter MRP1 (OR: 3.6, 95% CI: 1.6 – 8.4) and with the Val1188Glu-Cys1515Tyr (rs8187694-rs8187710) haplotype of the functionally similar MRP2 (OR: 2.3, 95% CI: 1.0 – 5.4). Polymorphisms in adrenergic receptors previously demonstrated to be predictive of heart failure were not associated with ACT.
The analysis was designed as a nested case-control study including all cases and matched controls of the cohort. Follow-up examinations were conducted every 3 months in the first 2 years and every 6 months thereafter. The follow-up examinations relevant to cardiotoxicity detection were scheduled at 1, 2 and 5 years after the therapy and included electrocardiography and echocardiography as well as physical examination.
The deviation of the genotype distributions from Hardy-Weinberg equilibrium (HWE) was tested with Pearson’s goodness-of-fit χ2 test. The lack of deviation of the genotype distribution among controls from HWE was necessary for the subsequent association testing. The latter was performed using the procedure by Freidlin, which is a modified Cochran-Armitage trend test, and by Fisher’s exact test. Results were also analyzed using multiple logistic regression for each genetic variant individually, adjusting for age, gender, total dose received, and for dosing interval (14 versus 21 days). In addition multiple logistic regression was used to investigate combinations of genetic variants that were individually significant. The significance level was set at 5%, as appropriate for screening purposes.
A total of 206 polymorphisms in 82 genes with conceivable role in ACT were genotyped. All but 17 variants were confirmed as biallelic markers, i.e. rs2032582 and rs746578 were triallelic, whereas 15 other variants were monomorphic. The genotypes of further 14 biallelic markers were not in Hardy-Weinberg equilibrium among controls. The remaining 175 variants from 73 genes were subsequently genotyped in cases and tested for associations. Fifty-nine out of 175 SNPs were in strong LD (r2>0.5) with at least one other SNP within the same gene. Using 116 independent SNPs (r2<0.5), we found no evidence for stratification of the patient population, as indicated by a variance inflation factor of 0.84.
The probability of clustering of 5 associations in 2 out of 10 functional groups of polymorphisms was calculated by simulations. The population stratification was investigated using genomic controls. Additional support for the associations found can be derived from the functional context of the findings.
| |
Figure 1: Odds ratios (OR) and confidence intervals (CI) to develop cardiotoxicity following doxorubicin treatment conferred by the predisposing alleles identified in this study. (Predisposing alleles not defined in this Figure.) |
Since in the study above a total of 206 polymorphisms in 82 genes have been genotyped as candidate gene polymorphisms in a case-control study an investigation of population stratification can be carried out in order to judge the possibility of spurious associations due to this stratification. The majority of these polymorphisms will not be involved with ACT so that they can be considered as markers for the assessment of population stratification. While we could find no evidence for population stratification in our nested case-control study we further investigated the impact of population stratification on case-control studies.
Methods accounting for population structure by using additional genetic markers broadly follow one of two concepts: Genomic Control (GC) and Structured Association (SA). GC tests empirically estimate the amount of overdispersion of the original test statistic. In the model based SA approach population structure is directly inferred and the test of association incorporates the estimated population structure. SA approaches themselves can be divided into one- and two-step approaches. The two-step approach consists of first modelling the population structure and then test for association based on this inferred structure. The one-step approach does this simultaneously. A remaining problem for SA is how to best estimate the number of underlying subpopulations.
We extended existing methods of SA and compared these to existing methods, as well as to GC in a large simulation study. We have shown that there are a lot of caveats, both for GC and SA, which should be taken into account, but that the investigation of population structure is indicated in realistic situations of large case-control studies with moderate population stratification.
Most importantly SA has to be applied with a clustering algorithm conditioning on the phenotype, otherwise a bias is introduced which leads to an inflated type-I error rate. This point has not been identified before as crucial. The approach of Satten et al. is a one-step approach which correctly conditions on the phenotype. It has the disadvantage that for each candidate locus the sample has to be clustered again. Thus, we have shown that it is sufficient to apply a two-step approach with the clustering algorithm conditioning on the phenotype. Compared to Satten et al. we do not lose other information than about the population substructure contained in the genotypes of the considered candidate locus.
For moderate population stratification a Wald test statistic should be preferred as a Structured Association test statistic in comparison to a likelihood ratio test. The main advantage of the Wald test is that allele frequency differences are averaged over subpopulations.
We also theoretically calculated the variation of the actual type-I error rate for the mean based GC test statistic and showed that this variation does not depend on the variance inflation factor itself, but only on the number of null loci. This is a weakness of GC compared to SA, where better clustering results are obtained with increasing FST. The fixation index FST measures the distance between the subpopulations with respect to the total population.
For method comparisons we simulated L marker loci for K discrete subpopulations in a case-control study of sample size N in each group and underlying population structure with fixation index FST, K=1-4, L=25-400, N=500-8000, FST=0.0025-0.04. These are realistic situations of large case-control studies with moderate population stratification. Relative Risks (RR) for the effect of the predisposing gene are simulated in such a manner that the prevalence is two-fold higher in subpopulation 2 than in subpopulation1, which can be expressed as RR=2 if subpopulation 2 is compared to subpopulation 1.
A disadvantage of GC turns out to be the large variation in estimating the variance inflation factor, as well as the power loss if population structure increases. We came to the overall conclusion that Structured Association, if applied correctly, is superior to GC, at least in the case of simple population structure as has been simulated here.
When applying our phenotype-dependent EM algorithm the correct number of subpopulations is inferred for almost all data sets and the median FST is quite accurately estimated with the basic parameter set of L=100, FST=0.01, N=2000, K=2, RR=2. The estimate of FST is substantially biased for smaller sample sizes N, due to the estimation method. For K=4 it is more difficult to infer the correct number of subpopulations.
Thus, in our study on doxorubicin-induced cardiotoxicity we decided to use the GC approach yielding no evidence for population stratification.
Another methodological aspect of the cardiotoxicity-study is the impact of multiple testing on a large variety of polymorphisms. The prospect of SNP-based genome-wide association analysis has been extensively discussed, but practical experiences remain limited. We performed an association study using a recently developed array of 11555 SNPs distributed throughout the human genome. 104 DNA samples using the NHL-B cohort were hybridized to these chips with an average call rate of 97% (range 85.3% – 98.6%). The resulting genome-wide scans were applied to distinguish between carriers and non-carriers of 37 test variants, used as surrogates for monogenic disease traits. The test variants were not contained in the chip. 55 test variants were selected arbitrarily from the databank of SNPs established for the NHL-B candidate SNP association project above. Only the 37 test variants with minor group frequency between 10% and 50% were selected, the 104 subjects were split into two groups according to the genotype with heterozygotes combined with the minor allele homozygotes.
Without adjustment 24% of the test variants were detected, but the positive predictive value was low (2%). Adjustment for multiple testing eliminated most false-positive associations, but the share of true positive associations decreased to 10-12%. We simulated fine-mapping of susceptibility loci by restricting testing to the immediate neighbourhood of test variants (+/- 5 Mb). This increased the proportion of correctly identified test variants to 22-27%. A bigenic inheritance reduced the sensitivity to 1%. Similarly adverse effect had reduction of allelic penetrance.
In summary, we demonstrate the feasibility and considerable specificity of SNP array-based association studies to detect variants underlying monogenic, highly penetrant traits. The outcome is affected by allelic frequencies of chip SNPs, by the ratio between simulated “cases” and “controls”, and by the degree of linkage disequilibrium. A major improvement is expected from raising the density of the SNP array.
We also aided in the statistical analysis evaluating the fidelity of genome-wide amplification of minute amounts of patient genomic DNA.
The elucidation of the genetic basis of anthracycline-induced cardiotoxicity as well as the genomic dissection of diastolic heart failure will be continued using the mutually complementary resources of clinical studies, mouse genetics, and molecular expression systems combined with our expertise in genetic epidemiological methods. In addition, we will continue to investigate methodological issues of direct concern of the studies on heart failure.
Work that has been already started is based on the dissection of heterogenous phenotypes and the investigation of longitudinal phenotypes with several follow-up points as in the supported heart failure projects.
Publications:
1. Wojnowski L et al. NAD(P)H oxidase and MRP genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Arch Pharmacol. 2004, 369(s1): R150.
2. Köhler and Bickeböller. Case-control association tests correcting for population stratification Ann Hum Genet. 2005, 69, 1-18.
3. Kulle et al. Application of genomewide SNP arrays for detection of simulated susceptibility loci. Hum Mutat. 2005, 25: 557-65. 4.
4. Tzvetkov et al. Genome-wide single-nucleotide polymorphism arrays demonstrate high fidelity of multiple displacement-based whole-genome amplification. Electrophoresis. 2005, 26: 710-715.
5. Bickeböller et al. Dissection of heterogenous phenotypes for quantitative trait mapping. Genetic Epidemiol. 2005 in press.
6. Rosenberger et al. Surrogate phenotype definition for alcohol use disorders: a genome-wide search for linkage and assoication. BMC Genetics. 2005, in press.
more information around the working group:
http://www.genepi.med.uni-goettingen.de/